Last update, Sept. 30, 2005
Publications on Virtual Endoscopy :
- Kaufman, A. et al. :
- Paik, D.S. et al. :
BibTeX references.
Penalized-Distance Volumetric Skeleton Algorithm
Ingmar Bitter,
Arie
Kaufman and Mie Sato
IEEE Transactions on Visualization and Computer Graphics, Vol.
7, No. 3,
pp. 195-206, July-Sept. 2001.
Abstract
This paper introduces a refined general
definition of a skeleton that
is based on a penalized-distance function and cannot create any of the
degenerate cases of the earlier Ceasar and Teasar algorithms.
Additionally, we provide an algorithm that finds the skeleton
accurately and rapidly. Our solution is fully automatic, which frees
the user from having to engage in manual data preprocessing. We present
the accurate skeletons computed on a number of test datasets. The
algorithm is very efficient as demonstrated by the running times which
were all below seven minutes.
Distance-Field Based
Skeletons for Virtual Navigation
Ming Wan, Frank Dachille and
Arie Kaufman
Proceedings of the conference on Visualization '01
pp. 239--246, San Diego, CA, USA, Oct. 2001.
Web site: http://www.cs.sunysb.edu/~vislab/projects/colonoscopy/
Abstract
We present a generic method
for rapid flight planning, virtual navigation and effective camera
control in a volumetric environment. Directly derived from an accurate
distance from boundary (DFB) field, our automatic path planning
algorithm rapidly generates centered flight paths, a skeleton, in the
navigable region of the virtual environment. Based on precomputed
flight paths and the DFB field, our dual-mode physically based camera
control model supports a smooth, safe, and sticking-free virtual
navigation with six degrees of freedom. By using these techniques,
combined with accelerated volume rendering, we have successfully
developed a real-time virtual colonoscopy system on low-cost PCs and
confirmed the high speed, high accuracy and robustness of our
techniques on more than 40 patient datasets.
Three-Dimensional
skeleton and centerline
generation based on an approximate minimum distance field
Yong Zhou (1)(2), Arie Kaufman
(2), Arthur W. Toga (1)
(1) Laboratory
of Neuro Imaging, UCLA School of Medicine, 710 Westwood Plz, RM
4-238 Reed, Los Angeles, CA 90024-1769, USA
(2) Center
for Visual Computing and Department of Computer Science, SUNY at
Stony Brook, Stony Brook, NY 11794-4400, USA E-mail: yzhou,
toga@loni.ucla.edu, ari@cs.sunysb.edu
The
Visual Computer, Volume 14 Issue 7 (1998) pp 303-314
Abstract
We propose an algorithm for generating 18-connected skeletons and
centerlines of 3D binary volume data sets. With of an approximate
minimum distance field, we express skeletons as a set of clusters with
a set of local maximum paths (LMpaths). Each cluster consists of
geometrically adjacent voxels with the same local maximum value.
Distinct clusters are connected by all possible LMpaths formed by local
maximum voxels snaking along, at most, three fixed directions until
they meet other clusters. As a 3D extension, we discuss an LMpath
traveling on a straight line before and after reaching a saddle point.
We generate the shortest centerline connecting two given points with
another similar minimum field over skeletal point sets. The results
generated by the algorithms on an experimental data set and colon CT
and brain MRI data sets demonstrate their efficiency.
Key words: 3D skeleton and centerline · Volume
visualization · Navigation · Distance transformation
Notes
Based on 3D-DT:
- Find local extrema clusters (in distance value)
- Link these cluster by a sort of 3D ridge following:
- uphill climbing from saddle points, defined as local minima
among skeletal points;
- "uphill climbing" is implemented as finding sets of local
maximum paths (LMpaths).
The particular 3D-DT used is very coarse: city-block-like version in 3D
(based on 6 direct ngbs. only: faces of voxels).
Automated
flight path planning for virtual endoscopy
Paik,
David S., Beaulieu,
Christopher F., Jeffrey, R. Brooke,
Rubin GD, Napel,
Sandy
Med Phys; 25(5):629-37, May 1998.
Abstract
In this paper, a novel technique for rapid and automatic computation
of flight paths for guiding virtual endoscopic exploration of 3D
medical images is described. While manually planning flight paths is a
tedious and time consuming task, our algorithm is automated and fast.
Our method for positioning the virtual camera is based on the medial
axis transform but is much more computationally efficient. By
iteratively correcting a path toward the medial axis, the necessity of
evaluating simple point criteria during morphological thinning is
eliminated. The virtual camera is also oriented in a stable viewing
direction, avoiding sudden twists and turns. We tested our algorithm on
volumetric data sets of eight colons, one aorta and one bronchial tree.
The algorithm computed the flight paths in several minutes per volume
on an inexpensive workstation with minimal computation time added for
multiple paths through branching structures (10%-13% per extra path).
The results of our algorithm are smooth, centralized paths that aid in
the task of navigation in virtual endoscopic exploration of
three-dimensional medical images.
 |
 |
|
Aorta & virtual camera pose along
the computed path.
|
One shot of the virtual angiography
sequence produced.
|
Notes
Previous approaches
Key framing :
- The user specifies manually the pose of a small subset of the
total number of frames to be rendered. Interpolating curves are fitted
to generate a continuous path and set of frames.
- Manual task very time consuming, prone to human errors.
Distance mapping :
- Comes from robotic path planning. A goal voxel
being selected, a distance transform ("map" in the text) is computed
from it within the medium of interest. A path is determined by
following the gradient of the DT.
- Tends to "hug the wall of the organ" (not central).
- Requires large amount of memeory to store the DT.
Iterative adjustment towards a central axis :
- Starting frame (2D slice) is chosen. Subsequent frames are
determined based on a 2D center of mass (in a plane perpendicular to
the previously determined position).
- Only approximates the 3D medial axis loci.
- Does not guarantee that the path will stay within the lumen
of the organ.
- No simple way of dealing with branching structure.
Thinning techniques to determine a medial axis :
- Iterative peeling off of voxels to determine a central 3D axis
structure (skeleton).
- Computationally expensive.
- Connectivity criteria is variable ("complex" in the text).
Proposed Path Planning Algorithm
Connectivity of voxels :
- 26-ngb for identified voxels (part of region/tissues of
interest).
- 6-ngb for nonidentified voxels (background).
- Surface voxels: identified voxels which have 6-ngb
connectivity with nonidentified voxels.
Segmentation :
- Region growing algorithm [Cline90].
- Hand corrections
Initial Path Selection :
- User specifies Start and End points: initial path members.
- A surface-based path (2D) is then determined between these 2
points.
Euclidean Distance Mapping (EDM):
- You start by assigning the start voxel a distance of 0. Then you
do a breadth-first search marking each voxel as the current
voxel's distance plus the Euclidean distance from the current voxel to
the voxel in consideration. Since voxels may be visited several times,
you only mark the distance if it is less than the current distance (or
hasn't been marked yet at all).
Thinning (based on EDM) :
- Step 1 of each iteration: Parallel "removal" (become
nonidentified) of all surface voxels (except path members) from the
structure of interest.
- Step 2 of each iteration: EDM computation:
- For the union of previous path voxels and actual surface
voxels.
- Step 3 of each iteration: Determine a new voxel path through the
EDM.
- The new path follows the new eroded surface whenever
possible.
- The new path traverses voxels of the old path only where
erosion has disconnected components of the surface.
- The previous 3 steps of thinning are repeated until the
structure of interest is thinned away and only the centralized path
remains.
Path sampling :
- The voxel path needs to be smoothed to void nonuiform camera
velocity.
Virtual Camera Orientation :
- Compute a viewing direction (look vector) that points toward
corners for the path ahead.
- Determine visibility by ray tracing
- Sequence of look vectors are smoothed.
- Camera roll (twist) is minimized frame-to-frame (through
projection in a picture plane).
 |
 |
|
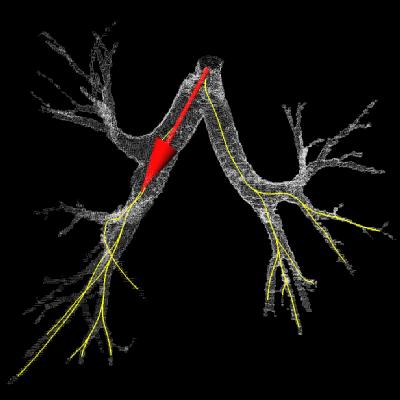
Bronchus & virtual camera pose
along the computed path.
|
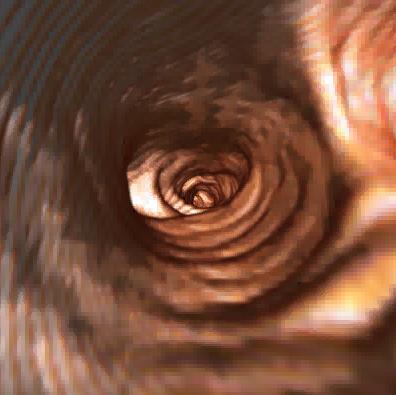
One shot of the virtual bronchoscopy
sequence produced.
|
 |
 |
|
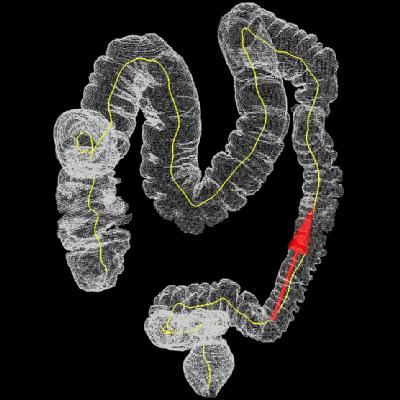
Colon & virtual camera pose along
the computed path.
|
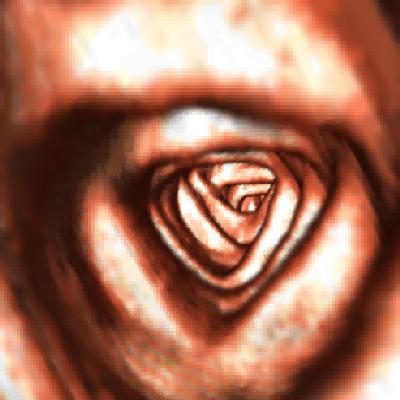
One shot of the virtual colonoscopy
sequence produced.
|
Page created & maintained by
Frederic Leymarie,
1998-2005.
Comments, suggestions, etc., mail to: ffl at gold dot ac dot uk







