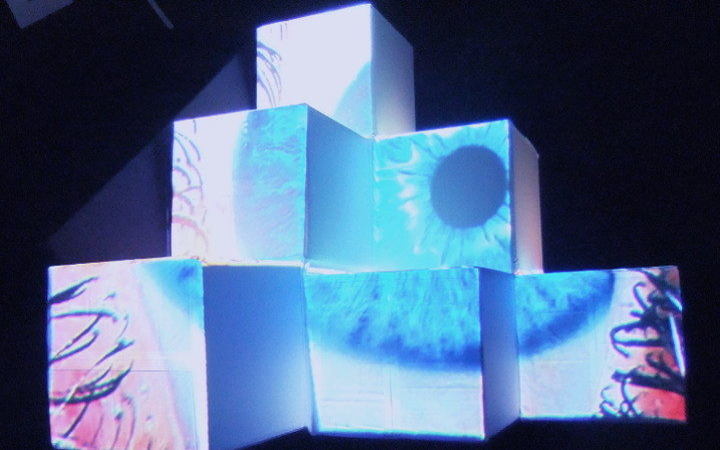
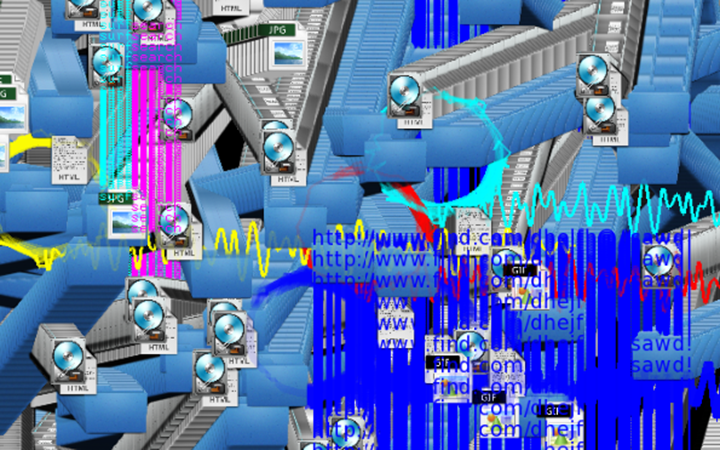
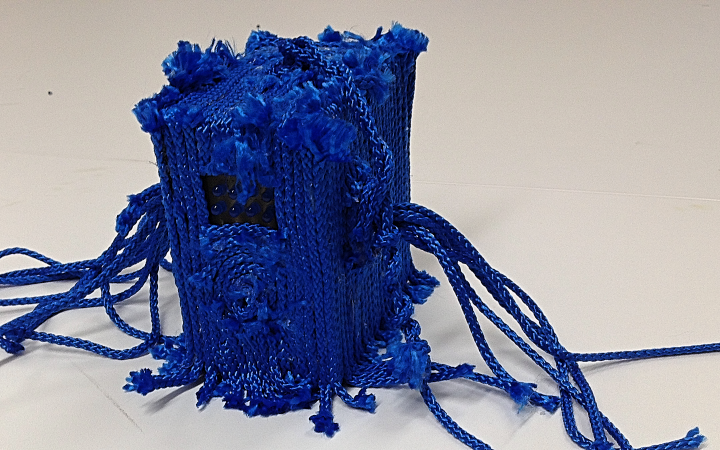
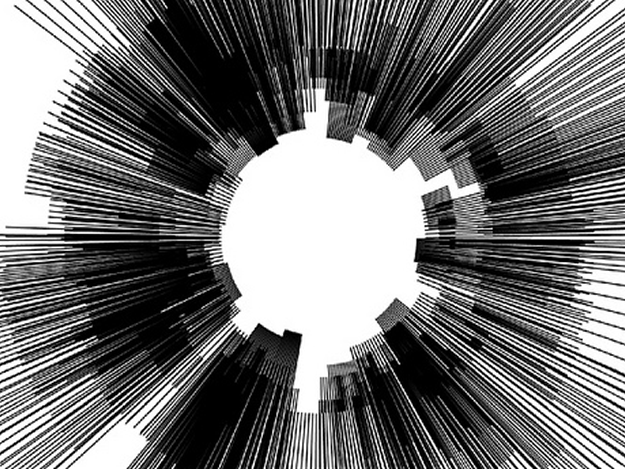
Degrees of Freedom
This generative work takes inspiration both from: a) electrophoresis; a technique used to estimate DNA's fragments length; and b) twin design; a methodological approach to estimate genetic and environmental sources of variation. It is about freedom and constrictions, and how they emerge from the relationship between coded information and complicated environments
produced by: Giacomo Bignardi
Music courtesy of Piet Row
Introduction
We are in the (post)genomic era. But what does it mean? And how can we interpret the results that are emerging from the genetic sciences? E.g., what does it mean to say that 30 to 60% of the variation in one's political ideologies can be explained by genetic factors (Hatemi et al. 2014); that polygenic risk scores might predict up to 11 percent of differences in educational attainment (Lee et al. 2018); or that music ability show substantially heritable components (Vinkhuyzen, Van der Sluis, Posthuma, & Boomsma, 2009)? One of the most common interpretations of genetic research is associated with the notion of determinism: it's pre-written, it's innate, it's fixed. But years of quantitative genetics research is suggesting the opposite. As (one of the two1) Robert Plomin stated in Blue Print(2018):
"Genetics is not a puppeteer pulling our strings. Genetic influences are probabilistic propensities...Genes are not destiny. You can change... Heritability describes what is but does not predict what could be"
There is no such thing as nature, genome or code without nurture, environment or physicality. It is not nature versus nurture, it is nature and nurture (Polderman et al. 2016). However, as a newbie researcher in the field, I was (and I still am) struggling to grasp what those interactions mean for the whole system (Populations) versus what they mean for the individual components of the same system (the person, a.k.a. you, me). So I played a little bit with the conceptual comparison between nature/genetic/code and nurture/environment/physicality. I let the generative piece inspire me. This is the result of that playing.
Concept and background research
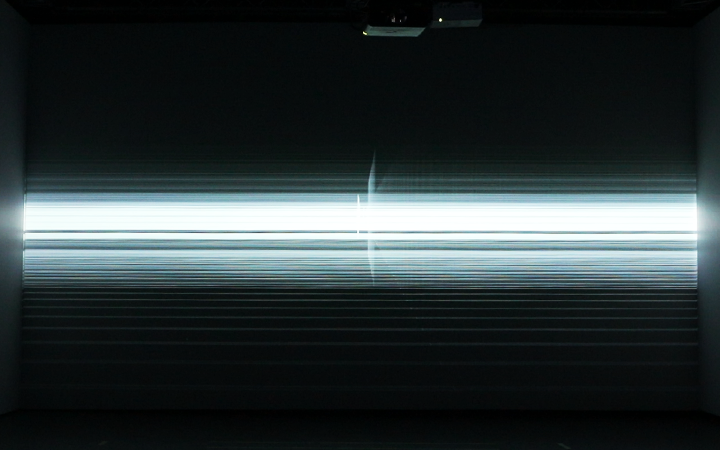
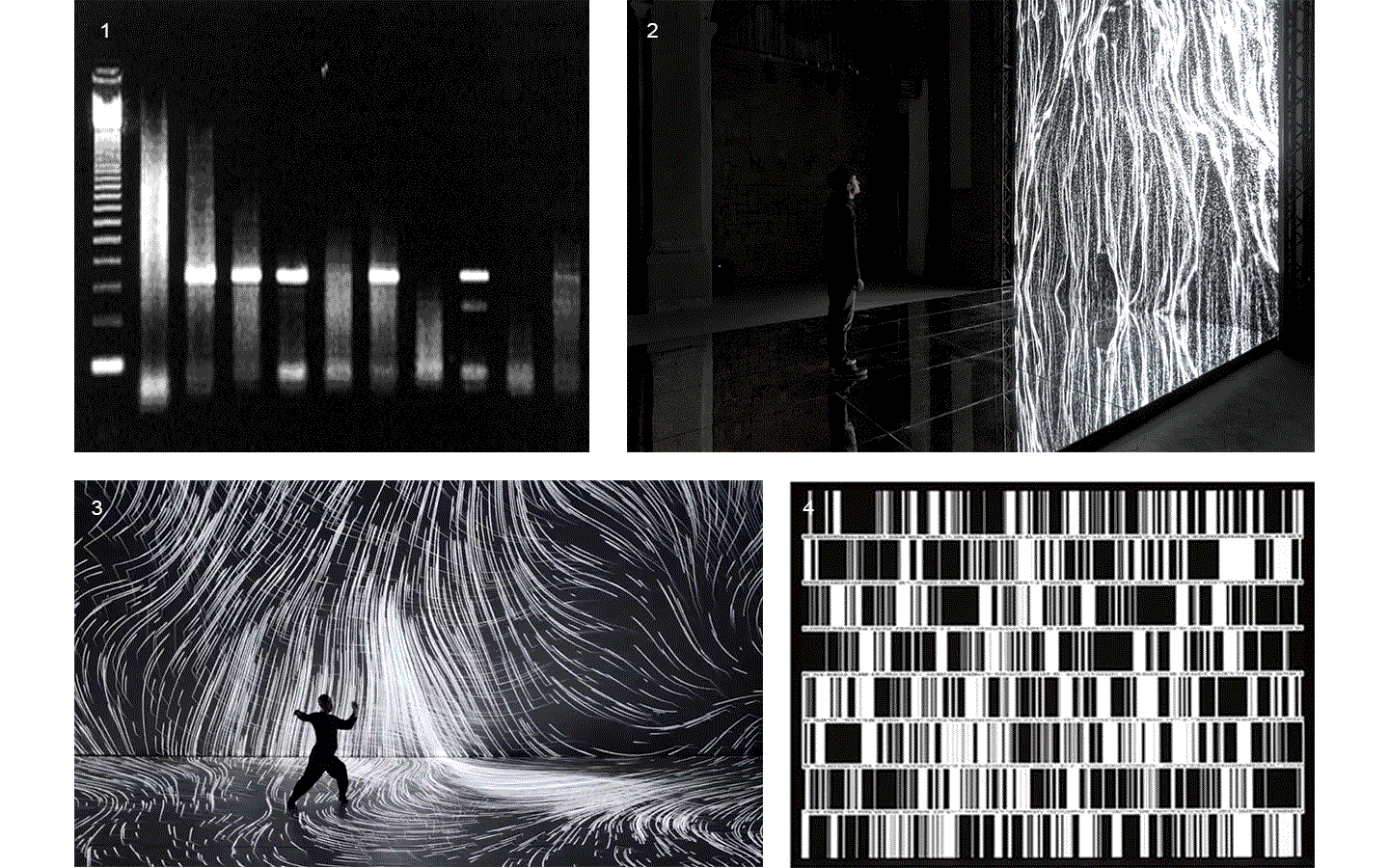
Electrophoresis iconography was used as the first visual source of inspiration. Briefly, Electrophoresis is used to assess the number of DNA base pairs (i.e, A-T, C-G) in chopped segments (visualized as rectangles) of DNA. The heavier the fragment is, the slower the fragment will travel through the gel2. This technique is used for example to detect sources of genetic differences between individuals at one or more specific locations of the genome. Ryoji Ikeda, consciously or unconsciously, visually displayed something similar in Datamatics (2006).
Moreover, the twin design was used as a source of conceptual inspiration. In a nutshell, twin designs are using identical and non-identical twins comparisons to make informed guesses on the role that nature and nurture play on specific traits (such as height for example).
Finally, the two biggest sources of visual inspirations were: 1) intensional practice performative work of Hiroaki Umeda (2015), and 2) Multiverse, a generative artwork from Fuse (2018).
1. Agarose gel electrophoresis: image from helicobacters present in gastric mucosa of human beings 2. Fuse, Multiverse (2018) 3. Hioraki Umeda, intensional practice (2015) 4. Ryoji Ikeda, Datamatics (2006)
Technical
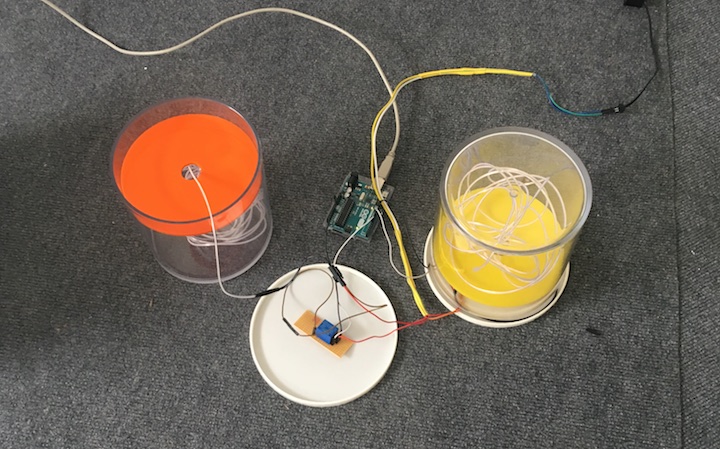
The entire piece was coded in openframeworks. ofxGui, ofxJSON, ofPimapper, ofXmlSettings addons were implemented to allow: 1) projection mapping; 2) compiling form a personal computer; 3) layering and timing management.
A MacBook Pro, 2,3 GHz Intel Core i5, macOS Hgh Sierra Version 10.13.6 was used as the machine where to run the code.
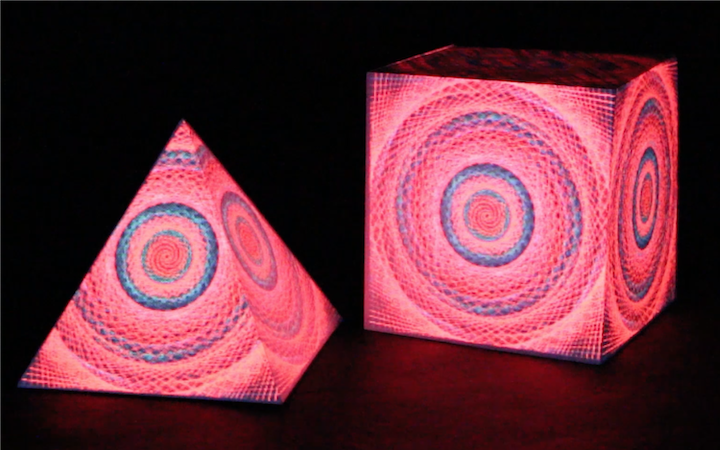
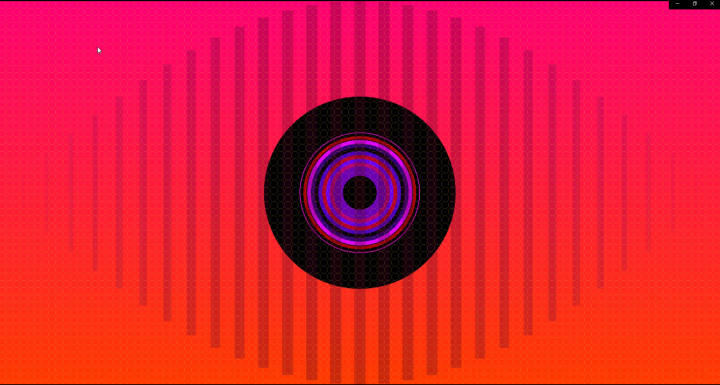
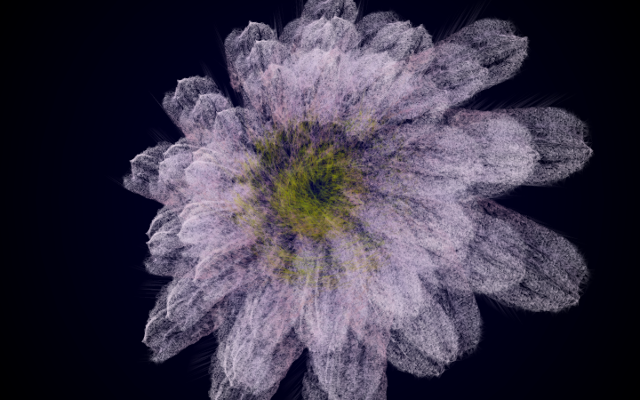
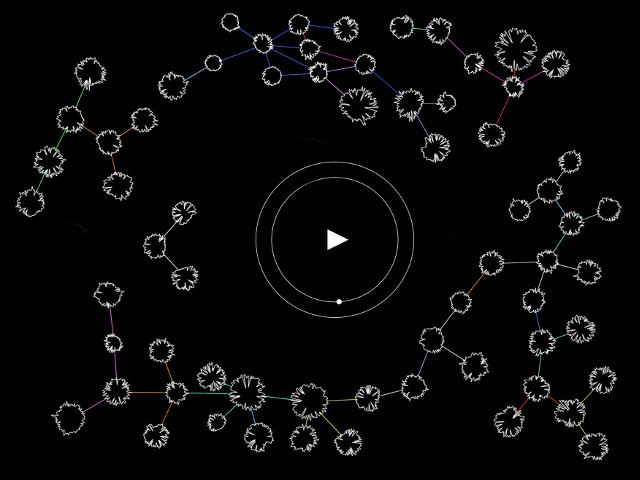
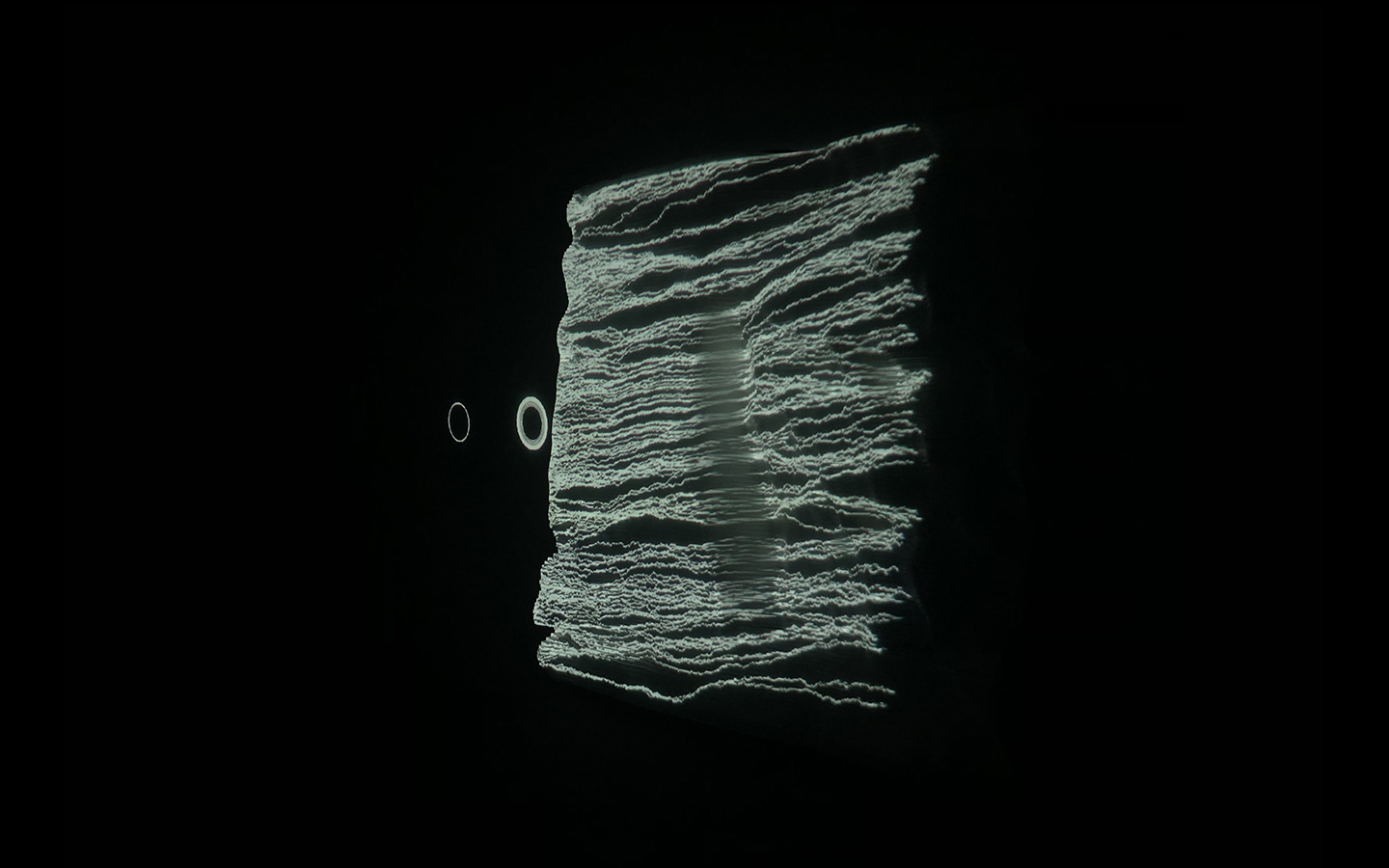
In this work, I just virtually coded identical twins, which are 100% genetically/code indistinguishable (almost). Then, the calculated distances between their DNA segments (simple rectangles) were mapped into radius lengths of circles. This allowed 'freedom' to be visually displayed.
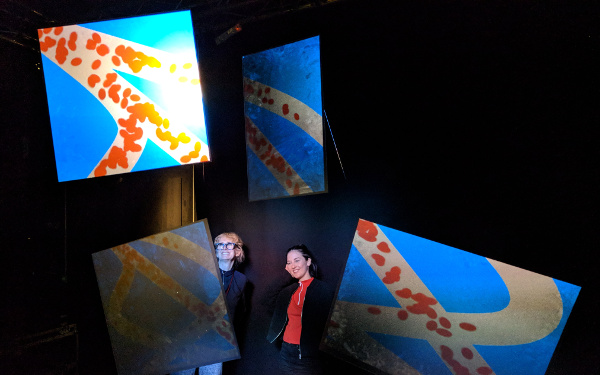
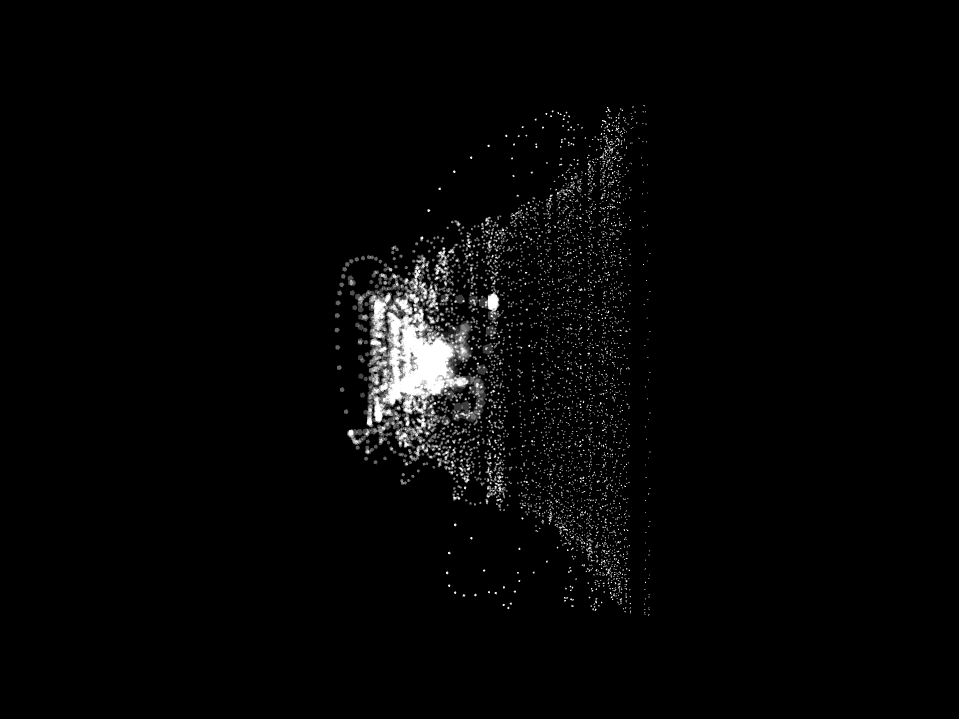
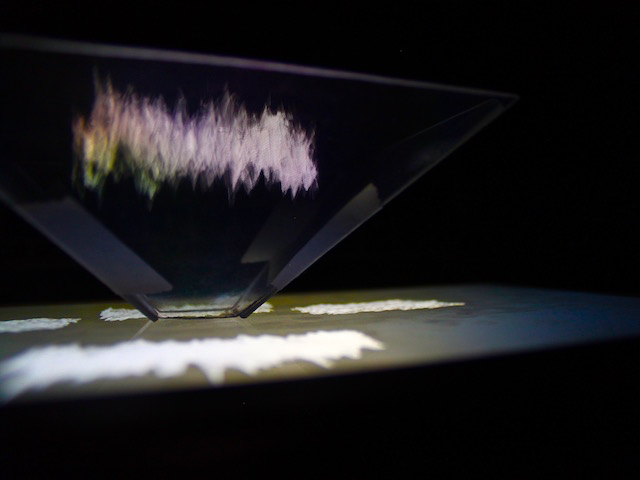
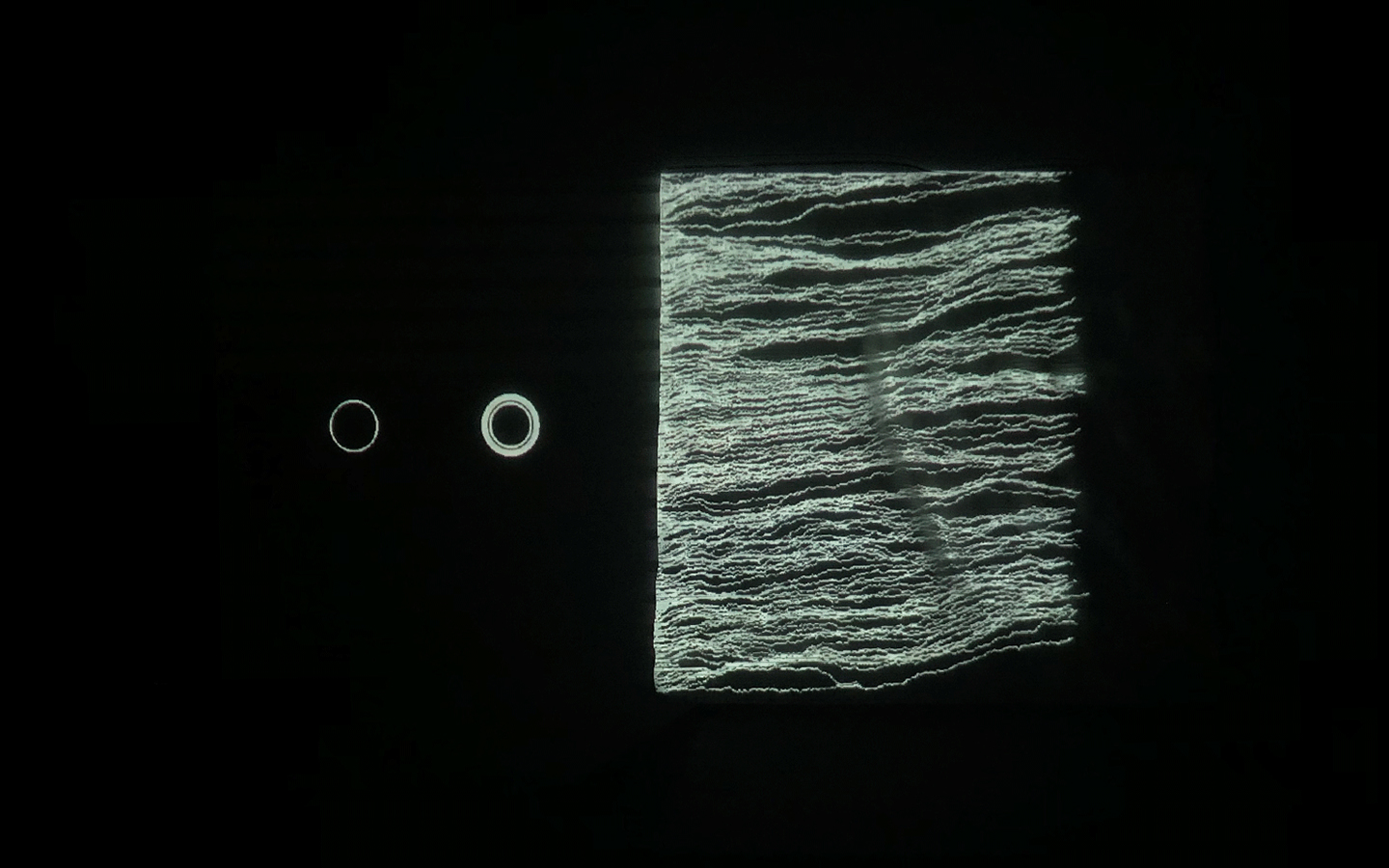
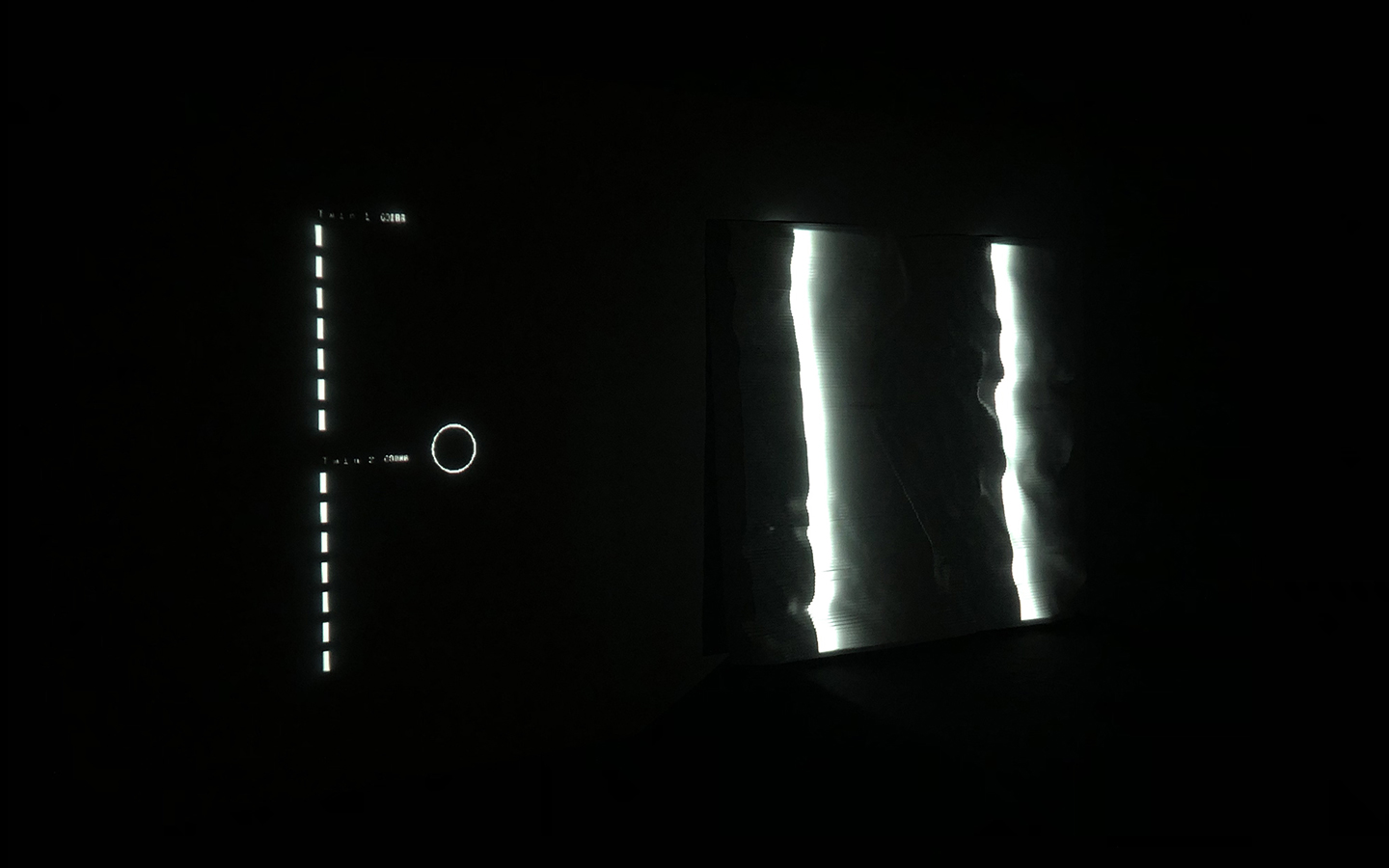
And there was shape3. At first, sinusoidal moving and flashing lights were introduced to scanning and showing the environment. Then the "DNA segments" were transformed into circles. n pixel of lights (the number of y pixel of the 2D model of the shape) started to interact with the shape, following a simple behavioral rule: to choose the next (up/up-right/right/down-right/down) brightest pixel (based on the 3D image), and to move. If only equally bright options were available, then the movements were randomly assigned. Again, the distances between the i and the i*n/2 pixels were computed and mapped into circles' radius. For comparison, the new circles generated on the shapes were displayed nearby the first ones. This is how the installation looked.
Self evaluation & Future development
There were a couple of things that did not work properly. First, I programmed the pixels' movements to restart from the horizontal coordinate zero when the width of the image was exceeded by it. However, the code behaved oddly and the visual result was a little bit unexpected. I decided to keep it, mostly due to its visual proprieties. Secondly, my first version of the code was crashing 1 out of 5 times. Adam He4 found a simple fix: to let the program know when to start to compute stuff from the image. That is, the image was properly loaded by simply adding 'if(imageName.isAllocated())' before the relevant code. The last issue was due to the edges of the object. Since ofPiMapper cannot take into account irregular 3D edges, the resulting mapping will never be perfect. So If you are thinking about doing something similar, keep that in mind.
It is quite hard to say whether I will continue to develop this project since my journey in this workshop has come to an end. However, I might implement some of the following in the later future: 1) Interaction (the real one), 2) More accurate genetic comparison, 3) different environments to see the shapes emerging from the collective behavior of the pixel vs. the degrees of freedom of the individual ones.
Footnotes
1. I am referring to a recent, amazing and funny article on Scientific American, by Scott Barry Kaufman. Surprised by some recent statement of Plomin, Kaufman replied … “there seems to be two different Plomins: (a) the careful, responsible scientist who did groundbreaking research in the field and provides the appropriate caveats, and (b) a publicly unleashed version of Plomin that says outrageous things that aren't even supported by his own research”. Don’t worry I cited version a.↩
2. If you are interested in the process of electrophoresis; 1) watch a quick video about electrophoresis (old fashion recording lecture); 2) understand the basic; 3) read some examples of applied electrophoresis; And 4) see the visual sources that inspire me.↩
3. The environment where the generative piece was project was modeled by Juliette Pépin.↩
4.Adam, you are a star. Thank you↩
References
Fuse (2018). Multiverse [Display]. Exhibited at Complesso Sant'Agostino - Chiesa di San Nicolò, Modena October 2009 – 7 March 2010. Retrieved from https://www.fuseworks.it/en/works/multiverse/
Lee, J. J., Wedow, R., Okbay, A., Kong, E., Maghzian, O., Zacher, M., ... & Fontana, M. A. (2018). Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nature genetics, 50(8), 1112. DOI: https://doi.org/10.1038/s41588-018-0147-3
Hatemi, P. K., Medland, S. E., Klemmensen, R., Oskarsson, S., Littvay, L., Dawes, C. T., ... & Christensen, K. (2014). Genetic influences on political ideologies: Twin analyses of 19 measures of political ideologies from five democracies and genome-wide findings from three populations. Behavior Genetics, 44(3), 282-294. DOI : 10.1007/s10519-014-9648-8
Humeda. H (2015).Intensional practice. Live performance,Modena, Italy 2018, November 17). Retrieved from http://hiroakiumeda.com/project.html#intensional
Ikeda, R. (2006), Datamatics. Retrieved from http://www.ryojiikeda.com/project/datamatics/
Plomin, R. (2018) Blueprint: How DNA Makes Us Who We Are. Retrieved from https://books.google.co.uk
Polderman, T. J., Benyamin, B., De Leeuw, C. A., Sullivan, P. F., Van Bochoven, A., Visscher, P. M., & Posthuma, D. (2015). Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nature genetics, 47(7), 702. DOI: https://doi.org/10.1038/ng.3285
Vinkhuyzen, A. A., Van der Sluis, S., Posthuma, D., & Boomsma, D. I. (2009). The heritability of aptitude and exceptional talent across different domains in adolescents and young adults. Behavior genetics, 39(4), 380-392. DOI: 10.1007/s10519-009-9260-5